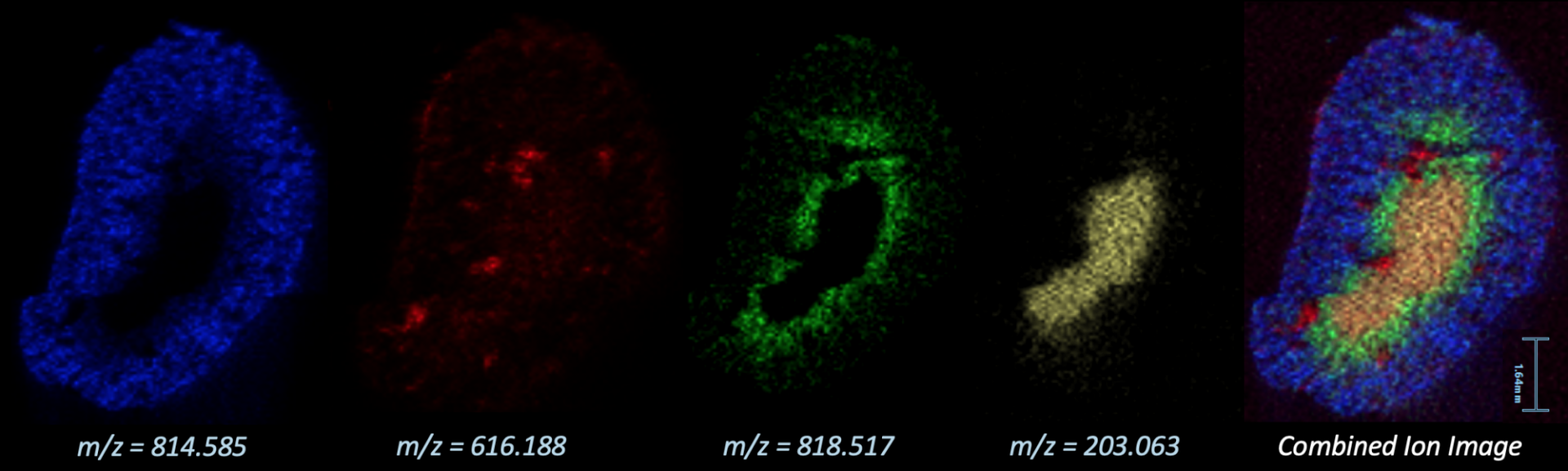
Mass Spec Imaging
Mass spec imaging (MSI) is a technique measuring chemical composition and linking it to spatial coordinates on a surface. The chemical composition is determined by mass spectrometry, which measures the mass-to-charge ratios (m/z's) of any ions that can be generated from the surface. Most commonly, the surface is a tissue section on a microscope slide; however, any flat surface could be analyzed given it has suitable dimensions and is properly prepared. The rest of this document will assume tissue imaging, but much of it could be applied to other surfaces as well.

Two instruments in the MSI facility are able to carry out imaging experiments. These include the Waters Synapt G2-Si QTOF for DESI (desorption electrospray ionization) imaging and the Bruker Ultraflextreme MALDI-TOF/TOF for MALDI (matrix-assisted laser desorption/ionization) imaging.
Waters Synapt G2-Si QTOF. The Synapt is a QTOF mass spectrometer that includes IMS (ion mobility spectrometry) for an added dimension of molecular specificity (i.e., collisional cross-section). Experiments with IMS included are dubbed HDMS in the Waters nomenclature. Fragmentation by CID (collision-induced dissociation) can be performed either before or after ion mobility separation to gain additional insight or specificity for a single ion (these are HDMS/MS experiments). The DESI imaging source has a maximum resolution of 50 um, while most experiments are carried out at 70-100 um. DESI is excellent for observing small molecules and metabolites (typically up to 1000 Da), and requires minimal sample preparation after sectioning onto a clean microscope slide.
Bruker Ultraflextreme MALDI-TOF/TOF. The Ultraflextreme is a TOF/TOF mass spectrometer that is has both MS and MS/MS capabilities. Fragmentation on this instrument relies on post-source decay. The MALDI imaging source has a laser shot frequency of 2 kHz with a maximum resolution of 20 um, and most experiments are carried out at 20-100 um. MALDI works well for small molecules, larger peptides, and even intact proteins with some optimization. This instrument requires that tissue sections are on slides with a clear, conductive coating (ITO), and the sample must be evenly sprayed with a MALDI matrix prior to analysis.
HTX TM Sprayer. This matrix sprayer allows for uniform and consistent coating of samples with a MALDI matrix. Methods can be optimized with respect to matrix concentration, solvent, solvent and nitrogen flow, and sprayer temperature, height, speed and direction. The sprayer can also apply a protease (e.g., trypsin) solution for on-tissue digestion.
Leica CM3050 S Cryomicrotome. This cryomicrotome allows users to bring frozen tissue for OCT-free cryosectioning onto slides for imaging.
Software. Waters' and Bruker's imaging software, HD Imaging and flexImaging, respectively, are available for image acquisition, viewing, and some analysis. There is a variety of free and commercial software that has been developed for advanced viewing and analysis of MS images, some of which are currently being tested in the facility. This will be updated when a software package is chosen for purchase.